Page 11 - iCC044-201501_March
P. 11
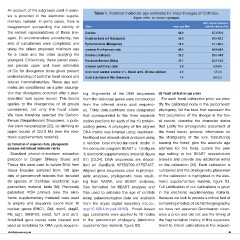
An account of the outgroups used in analy- Table 1. Posterior molecular age estimates for major lineages of Cichlidae.
sis is provided in the electronic supple- Ages refer to crown groups.
mentary material. In some cases, there is
disagreement surrounding the identity of Clade mean age (Ma) 95% highest posterior
the earliest representatives of these line- density interval (Ma)
ages. To accommodate uncertainty, two
sets of calculations were completed: one Cichlidae 64.9 57.3-76.0
using the oldest proposed minimum age Etroplinae (India and Madagascar) 36.0 30.3-42.2
for a clade and the other applying the Ptychochrominae (Madagascar) 38.2 31.7-46.4
youngest. Collectively, these paired analy- unnamed Afro-American clade 46.4 40.9-54.9
ses provide upper and lower estimates Cichlinae (neotropics) 29.2 25.5-34.8
of CIs for divergence times given present Pseudocrenilabrinae (Africa) 43.7 38.2-51.6
understanding of both the fossil record and unnamed east African clade 8.0 6.9-9.5
teleost interrelationships. These age esti- most recent common ancestor of L. Malawi and L. Victoria radiations 2.3 1.7-3.1
mates are conditioned on a prior assump- Crater Lake Barombi Mbo (Cameroon) 1.4 0.8-2.3
tion that divergence occurred after a user-
specified hard upper bound. This bound ing. Alignments of the DNA sequences (d) Fossil calibration age priors
applies to the divergences of all groups from the individual genes were constructed For each fossil calibration prior, we iden-
considered, not only the focal clade. from the inferred amino acid sequenc-
We have therefore selected the Carbon- es. Thirty data partitions were designated tify the calibrated node in the percomorph
iferous (Serpukhovian) Discoserra, a puta- that corresponded to the three separate phylogeny, list the taxa that represent the
tive stem neopterygian [22], as defining an codon positions for each of the 10 protein- first occurrence of the lineage in the fos-
upper bound of 322.8 Ma (see the elec- coding genes. A phylogeny of the aligned sil record, describe the character states
tronic supplementary material). DNA matrix was inferred using maximum- that justify the phylogenetic placement of
likelihood and relaxed-clock analyses using the fossil taxon, provide information on
(c) Collection of sequence data, phylogenetic a random local molecular-clock model in the stratigraphy of the rock formation(s)
analyses and relaxed molecular clocks the computer program BEAST v. 1.6 (figure bearing the fossil, give the absolute age
1; electronic supplementary material, figure estimate for the fossil, outline the prior
Standard phenol-chloroform extraction S1) [23,24]. DNA sequences are depos- age setting in the BEAST relaxed-clock
protocol or Qiagen DNeasy Blood and ited on GenBank KF556709-KF557487. analysis and provide any additional notes
Tissue kits were used to isolate DNA from Aligned gene sequences used in phyloge- on the calibration [25]. Each calibration is
tissue biopsies sampled from 158 spe- netic analyses, phylogenetic trees result- numbered and the phylogenetic placement
cies of percomorph teleosts that included ing from RAXML and BEAST analyses, of the calibration is highlighted in the elec-
89 species of Cichlidae (electronic sup- files formatted for BEAST analyses and tronic supplementary material, figure S3.
plementary material, table S6). Previously files used to estimate the age of cichlids Full justification of our calibrations is given
published PCR primers (see the elec- using palaeontological data are available in the electronic supplementary material.
tronic supplementary material) were used from the dryad digital repository (dx.doi. Because we look to provide a critical test of
to amplify and sequence exons from 10 org/10.5061/dryad.48f62). Fossil-based competing models of cichlid biogeography,
nuclear genes (ENC1, Glyt, myh6, plagl2, age constraints were applied to 10 nodes we have not assumed Gondwanan vicari-
Ptr, rag1, SH3PX3, sreb2, tbr1 and zic1). in the percomorph phylogeny (electronic ance a priori and did not use the timing of
Amplified gene copies were cleaned and supplementary material, figure S2). the fragmentation history of this supercon-
used as templates for DNA cycle sequenc- tinent to inform calibrations in the relaxed-
cichlidpower.org.au 9